This view provides the user interface to extract the peaks of tensors and the spherical harmonic representation of Q-Balls.
Available sections:
Input Data
Mandatory Input:
- DTI image or image containing the spherical harmonic coefficients. The SH coefficient images can be obtain from the Q-Ball reconstruction view by enabling the checkbox in the advanced options.
Optional Input:
- Binary mask to define the extraction area.
Output Data
- Vector field: 3D representation of the resulting peaks. Only for visualization purposes (the peaks are scaled additionally to the specified normalization to improve the visualization)!
- # Directions per Voxel: Image containing the number of extracted peaks per voxel as image value.
- Direction Images: One image for each of the extracted peaks per voxel. Each voxel contains one direction vector as image value. Use this output for evaluation purposes of the extracted peaks.
Peak Extraction Methods
- If a tensor image is used as input, the output is simply the largest eigenvector of each voxel.
- The finite differences extraction uses a higly sampled version of the image ODFs, extracts all local maxima and clusters the resulting directions that point in a similar direction.
- For details about the analytical method (experimental) please refer to [1].
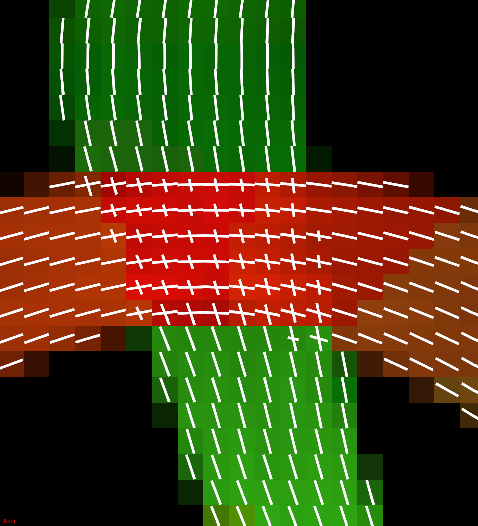
Peaks of a fiber crossing extracted using finite differences method.
Input Parameters
- Vector normalization method (no normalization, maximum normalization of the vecors of one voxel and independent normalization of each vecor).
- SH Order: Specify the order of the spherical harmonic coefficients.
- Maximum number of peaks to extract. If more peaks are found only the largest are kept.
- Threshold to discard small peaks. Value relative to the largest peak of the respective voxel.
- Absolute threshold on the peak size. To evaluate this threshold the peaks are additionally weighted by their GFA (low GFA voxels are more likely to be discarded). This threshold is only used for the finite differences extraction method.
References
[1] Aganj et al. Proceedings of the Thirteenth International Conference on Medical Image Computing and Computer Assisted Intervention 2010
