Table of Contents
Disclaimer
- The Segmentation Task List View and MITK Segmentation Task Lists in general are still in an experimental stage.
- While we try to minimize breaking changes in the future, we cannot give any guarantees at the moment.
- We strongly advise to ignore the Data Manager regarding loading and unloading of data while using the Segmentation Task List View, as it will completely take over these operations and may run into invalid states otherwise.
Overview
As the Segmentation Task List View is typically used in combination with the Segmentation View, we suggest to move the Segmentatiom View to the other side of the application (e.g. on top of the Data Manager), to see both views at the same time.
To unlock the Segmentation Task List View, unload everything but a single MITK Segmentation Task List. The remaining Segmentation Task List will be automatically selected.
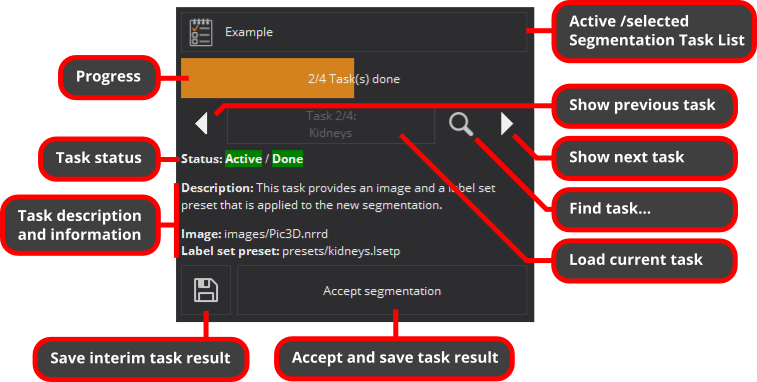
The Segmentation Task List View shows the progress of the whole Segmentation Task List, e.g., the number of the tasks marked as done vs. the total number of available tasks.
Below the progress indictator you can navigate between tasks, read their descriptions and related information, as well as load/activate the currently shown task. This will unload all data from a previously active task, if any, and load all data of the current task. To prevent any accidental data loss, unsaved task data will interfere task switches and you can decide on how to proceed.
Above the task description, the status of tasks is displayed as a pair of colored labels, indicating if a task is either active or inactive and if it is considered not to be done, having unsaved changes, or to be done.
With the bottom two buttons you can either save an interim result (the task is considered not to be done), or accept and save the task result. You can still edit accepted tasks but in contrast to interim results, the task is considered to be done.
Keyboard shortcuts
The Segmentation Task List View can be used even more efficiently with the following set of keyboard shortcuts:
Ctrl+Alt+P: Navigate to previous taskCtrl+Shift+P: Navigate to previous undone task (orShift+ click on resp. button)Ctrl+Alt+N: Navigate to next taskCtrl+Shift+N: Navigate to next undone task (orShift+ click on resp. button)Ctrl+Alt+L: Load currently shown taskCtrl+Alt+S: Store interim resultCtrl+Alt+A: Accept task and store resultCtrl+F: Find task...
Finding tasks
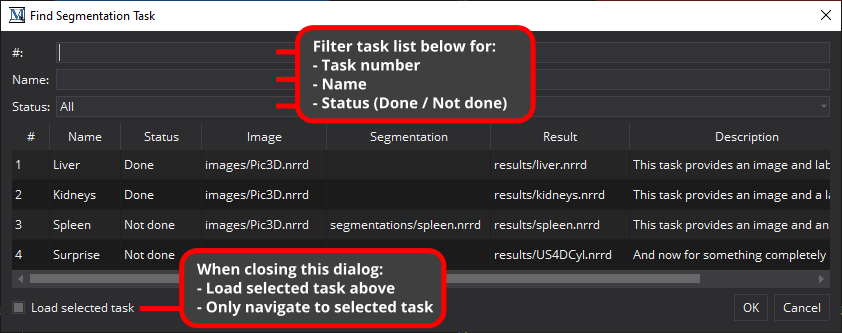
To find a specific task in a segmentation task list, click on the "Find task..." button (magnifier icon) or press Ctrl + F. A dedicated dialog will appear for filtering and searching the current segmentation task list. Enter a task number and press Return to immediately load the according task or filter the shown list of tasks and double click on a result to load the task.
MITK Segmentation Task List file format
MITK Segmentation Task List files are JSON files containing a JSON object as root. It must contain the two mandatory properties FileFormat and Version:
We also recommend to specify an optional Name that is used in the application if present instead of the plain filename of the JSON file:
Tasks
The root object must also contain a mandatory Tasks array, containing JSON objects that specify the individual tasks of the task list. A minimum task object must contain Image and Result file paths. Image refers to the patient image and Result refers to the path were the resulting segmentation is expected to be stored. Paths can be absolute or relative to the JSON file.
In addition, tasks can contain a bunch of optional properties that mainly specify a segmentation a user starts with:
Name(string): A name for the task.Description(string): A short description/definition of the task.LabelName(string): The name of the first label in a new segmentation that is created for the task on the fly.LabelNameSuggestions(file path): A Label Suggestions JSON file (example in next comment) specifying names and optional colors, that are suggested to the user for new labels in the segmentation.Preset(file path): A Label Set Preset XML file in MITK's .lsetp file format. The preset is applied to a new segmentation that is created for the task on the fly. We recommend to use the Segmentation plugin of the MITK Workbench to create such label set preset files as described in its {key F1} user guide.Segmentation(file path): A pre-segmentation that a user can start with or has to refine for example.Dynamic(boolean): In caseImagerefers to a dynamic (3d+t) image, specifies whether the segmentation should be static (false), i.e. equal for all time steps, or dynamic (true), i.e. individual for each time step.
Task defaults / common properties
If a task list contains multiple tasks with common properties, they do not have to be specified for each and every task again and again. Instead, the root object can contain an optional Defaults object that is identical in format to the tasks specified above. As the name indicates, default properties can still be overridden by individual tasks if they are specified explicitly.
There is a single exception, though: A Defaults object must not contain a Result file path, since result files of tasks must be distinct by definition.
Example
The following example is a complete showcase of the properties and features listed above. It specifies 4 tasks. 3 tasks refer to the same patient image so it is specified as default.
Remember that the only task property required to be distinct is Result so you are pretty free in your task design. For simplicity, we chose to define tasks around organs for this example and named the tasks accordingly:
MITK Label Suggestions file format
The Label Suggestions JSON file format mentioned above to specify a list of suggested names and optional colors for new labels is as follows:
