the data
The first step we take is to resample the data. To do so, we open the "Radiomics Transformation" View and select the "Resample Image" panel. We start by resampling the original image and therefore select the original picture (for us, Pic3D). Right-clicking on the image in the "Data Manager" and selecting the option "Details" gives us more information on the image. As we can see, our image has a spacing of [1, 1, 3], with an inplane resolution of 1x1mm and a out-of-plane resolution of 3 mm. We therefore decide to resample the image to an isotropic resolution of 1x1x1 mm.
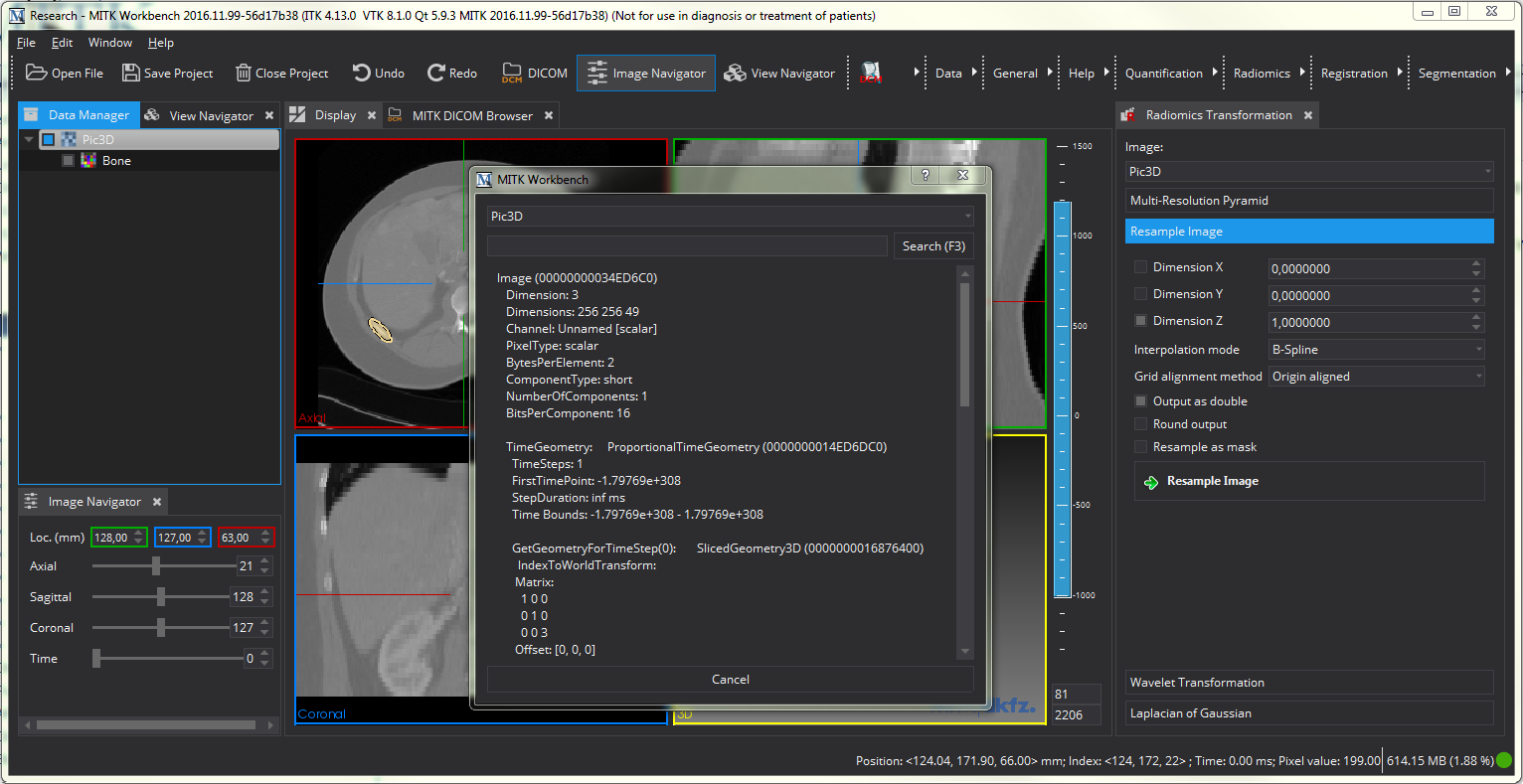
To resample the image, we de-select "Dimension X" and "Dimension Y" option and set the "Dimension Z" option to 1, as indiciated by the image above. This tells the resampling algorithm to change only the last dimension to the value we specified. We further select to have the output image as double and chose B-Spline as resampling algorithm. This is a fast and still accurate option for resampling. To learn more about the other interpolation modes, refer to The MatchPoint Image Mapper View .
After resampling the original image, we also need to resample the segmentation. For this, we select the segmentation, leave the dimensions unchanged. Remove the "Output as double" option, as segmentations are not double values and choose a linear interpolation, which seems to be a better solution for resampling masks. We also check the option that we are resampling a mask.
After performing those two steps, there should be two additional, resampled images in the "Data Manager".
As a second step, we calculate some Laplacian of Gaussian images of the resampled image that allow us to capture more detailed information. For this, we select the panel "Laplacian of Gaussian" of the "Radiomics Transformation"-view and perform the algorithm three times with different sigma values (we chose 1,2, and 4). Make sure that you selected the right image to calculate the image, i.e. the resampled image.
Finally, we clear the mask to obtain a clear segmentation of the target structure and remove possible resampling artifacts. To do so, we open the "Radiomics Mask Processing" View and select the resampled image and the resampled mask. We then select a lower limit only and set it to 160. Since we are working with MR, this is not a fixed value but something we manually determined. With this set, we perform the mask reducing by cklicking "Clean Mask based on Intervall". After this, we have the resampled image, three LoG images and a resampled and cleaned mask. The result should look similar to the next picture. You can also see the final image structure we obtained from our processing. It might help you to compare your results, although it is not necessary to obtain the same structure as long as you have all necessary images.

