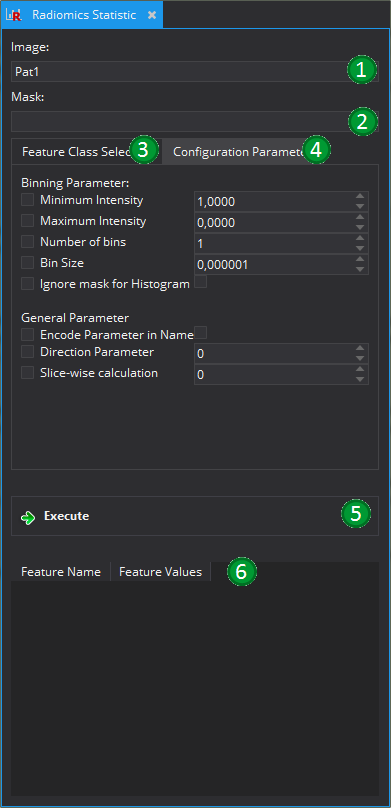
This is a basic view to actually calculate different image features, for example those necessary for radiomic studies. It is possible to select the required feature classes that should be calculated from the given image.
In order to be able to calculate the features it is necessary to define an image (1) and a corresponding mask (2). The image will be converted to an double image during the feature calculation and the mask must be a an Multi-Level Segmentation. If the mask is a standard mask, it is possible to convert the mask into a multilabel mask by right-clicking on the corresponding image on the "Data Manager" and selecting the point "Convert to Segmentation". The image and the segmentation must have the same geometry, e.g. the same spacing, origin, transformation matrix, and same dimensions.
Before calculating the features, it is necessary to select the feature classes that should be calculated in the corresponding panel (3). Each feature class consists of a set of Radiomics Features that share common proporties in the calculation process. All possible feature classes are derived from the AbstractGlobalImageFeature class. To get more details about the individual features, please see the corresponding classes.
It is possible to configure all feature classes within the "Feature Class Selection"-panel (3) but it is also possible to set more global configuration parameters in teh "Configuration Parameter" panel (4). The options set here will influence all features if the corresponding parameter is not explicitly overwritten. For a detailed description of the parameters, see the corresponding subsection.
After starting the calculation process with the "Execute" Button (5), the features will be calculated and the result shown in the result table (6) at the end of the view.
Parameters
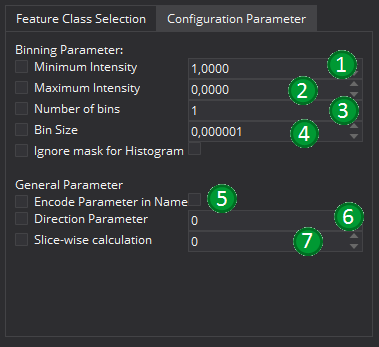
There are two types of possible parameters. The Binning Parameters (1-4) which helps to control the histogram building process that most feature classes use. The second group represent more general parameter (5-7). All parameters are disabled by default (Checkbox left of feature description text), as there is no default value that fits all cases. In order to set a parameter, it must be first enabled. This is especially important for binary options like ignorign the mask for the histogram.
The histogram options allow to define the histogram that is used for many feature classes. It is possible to define a minimum (1) and a maximum (2) intensity value for the histogram. In addition to this there is the possiblility to set the number of bins (3) or the bin size (4) for each histogram. A detailed description of each parameter is given in the description of the AbstractGlobalImageFeature class.
With "Encode Parameter in Name" (5) it is possible to allow the actual parameter set to be encoded in the final feature name. By default, the resulting feature names will only contain the feature class and the actual feature name. If this option is checked, also all relevant parameters are encoded in the name of each feature.
The parameter "Direction Parameter" allows to restrict the calculation of the feature to a given dimension, if this is supported by the feature classes. If it is set to 0, all directions are considered. If it is set to 1, only one direction is considered (mainly for testing purpose). If it is set to values 2 to n, the value-2 dimension is not considered during the feature calculation.
The parameter "Slice-wise calculation" allows to perform slice-wise calculations. Generally, it can be used with all parameters but not all features are well-defined for 2D data (for example, volume based features expect at least 3 dimensions). The value defied here gives the axis along which the split is performed.